
EXOSOME ISOLATION (PLASMA, URINE)
SubXTM-Exo kit is designed for isolation of exosomes from liquid biopsies (plasma, serum, etc.). Our proprietary bi-functional substance (SubXTM) interacts with exosomes via phospholipids. Each molecule of SubXTM can anchor two exosomes and thus produce dimers and then oligomers of the extracellular vesicles. SubXTM can oligomerize exosomes in both highly proteinaceous bioliquids (serum, plasma) and in high salt biofluids (urine, tears, etc.). Excess of SubXTM results in oligomerization of up to 10-15 exosomes and formation of micron-size particles that are easily precipitated in a brief 14K x g centrifugation step. A specially designed buffer allows for reconstitution of the pelleted exosomes back to free monomer state suitable for downstream applications.
SubXTM does not bind free phosphoproteins and phospholipids or its aqueous micelles (i.e. not inegrated into membranes).
See supporting experimantal data here.
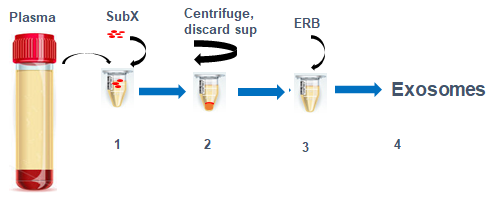
Figure 1. Exosome isolation steps. All procedures after mixing plasma or urine with SubXTM are done in one tube. Exosomes are recovered from SubXTM-Exosomes pellet by reconstitution in a special buffer ERB.
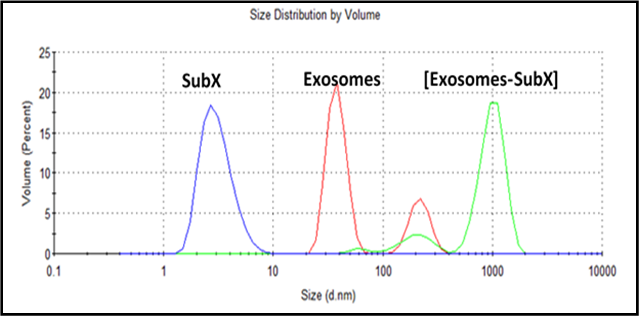
Figure 2. SubXTM binding with exosomes in urine. SubXTM particles size is around 3 nm (blue line). Urine (5,000 rpm supernatant) exosomes have a bimodal distribution of 40 nm peak and 220 nm peak (red line). [Urine (100ul) + SubXTM (20ul)] suspension (green) also have a (shifted) bimodal distribution of minor 210 nm peak and main 1000 nm peak of particle aggregates.
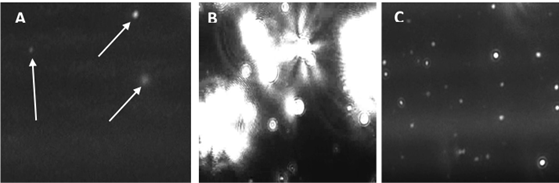
Figure 3. Snapshot images of exosomes taken from NanoSight NS300 videos. A - exosomes in urine; B –SubXTMoligomerizes exosomes to form large complexes with size range >400 - 1200 nm; C – exosomes reconstituted in ERB solution, size within the range 50-120nm
To see video please click the button below:

Figure 4. The characteristic cup shape of the exosomes is due to the static force on the grid disc that flattens the exosome shape. The cup and edges of the exosome take up the Uranyl Acetate which facilitates imaging of the exosomes under TEM electron beams. The sample used for this image was urine from patient with ovarian cancer. Image is courtesy of Dr. Anna Lokshin (University of Pittsburgh Cancer Institute).
References:
Shtam T, et al. (2020) Evaluation of immune and chemical precipitation methods for plasma exosome isolation. PLOS ONE 15(11): e0242732. https://doi.org/10.1371/journal.pone.0242732

