
LIQUID BIOPSIES, EXOSOMES, RNA AND DNA ISOLATION
CBI offers prospective collection of plasma samples from patients, kits and reagents for efficient isolation of circulating exosomes and cell-free DNA (cfDNA) from liquid biopsies. Our SubXTM cfDNA isolation kits have been specifically developed for efficient recovery of small DNA fragments (<35bp). SubXTM-Exo-DNA kit is designed for simultaneous isolation of exosomes and cfDNA directly from liquid biopsies (plasma or serum) without Proteinase K.
- Preparations of exosomes and circulating cell-free DNA derived from bioliquids employing SubXTM technology are not contaminated with free phosphoproteins, phospholipids or its aqueous micelles.
- SubXTM technology allows for separation of extracellular cfDNA and exosomes in a single protocol without cross-contamination.
Introducing new SubXTM-RNA kit for isolation viral RNA!!!
SubXTM Plasma cfDNA and SubXTM-Exo-DNA Isolation Kits - Liquid Biopsy Made Easy
 Unique features of SubXTM technology:
Unique features of SubXTM technology:
i - SubXTM binds DNA under physiological conditions (e.g. directly in biological liquids) with high affinity degree, followed by adsorption of the [DNA-SubXTM] complex on a solid phase matrix. SubXTM captures DNA via its phosphate groups and allows for elimination of bias related to both AT/GC content and DNA fragments length, thus improving extraction efficacy and accuracy of downstream applications. Since our method does not require chaotropic salts for DNA capture, no dilution of starting material (and cfDNA) will take place.
ii - SubXTM captures DNA in highly proteinaceous solutions without Proteinase K employment since it can dissociate or squeeze out histones and other DNA binding proteins from the DNA-Protein nucleosome complex.
iii - Both ends of SubXTM molecule display phospholipid clusters binding groups. This feature allows each molecule of SubXTM to anchor two exosomes (i.e. dimerize) and since several SubXTM molecules can bind single exosome this results in further oligomerization.
iv - SubXTM oligomerizes exosomes in both highly proteinaceous bioliquids (serum, plasma) and in high salt biofluids (urine, tears, etc.). Excess of SubXTM molecules in the solution results in oligomerization of up to 10-20 exosomes and formation of micron-size particles that are easily precipitated in a brief 14K x g centrifugation step. A specially designed buffer allows for reconstitution of the pelleted exosomes back to free monomer format suitable for downstream applications.
v – Different degrees of affinity of SubXTM to DNA (strong) and to exosome phospholipid complexes (mild) allow to separate exosomes and cfDNA from the total pellet. Exosomes are easily recovered in monomer format by special low salt reconstitution buffer while tightly bound [SubXTM-cfDNA-beads] complex remains in pellet and cfDNA further isolated by special solutions.
vi – SubXTM does not bind free phosphoproteins and phospholipids or its aqueous micelles (i.e. not inegrated into membranes). To anchor target molecule/microvesicle SubXTM requires oligophosphate cluster of at least 20 residues. For example, casein contains cluster of 4 phosphoserines, which is not not enough to form casein-SubXTM complex. Micelles formed by ovalbumin can mimic extracellular vesicles by the size distribution ranging from of 100 nm to 250 nm. Addition of SubXTM also does not results in micelles aggregation, instead dissociation of aggregates back to 2-3 nm particles occurs. See supporting experimantal data below.
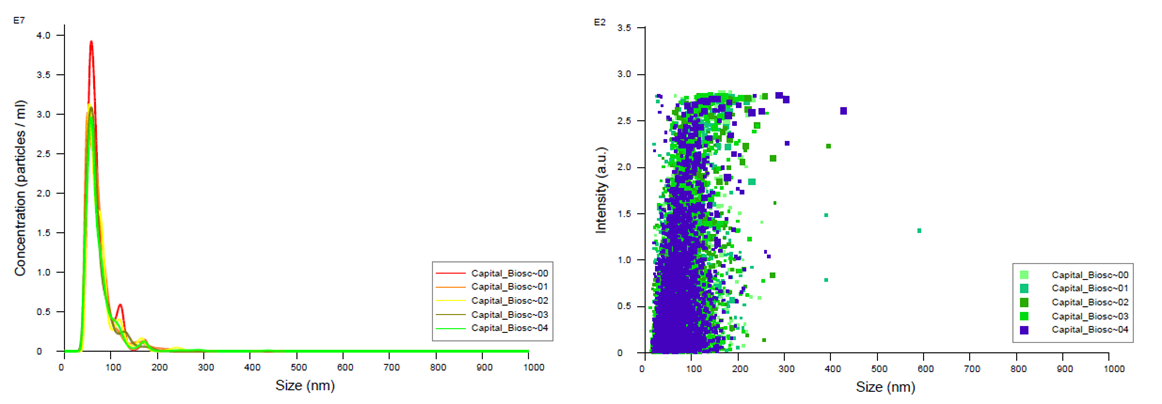
Figure 1. FTLA Concentration / Size and Intensity / Size graphs for exosomes isolated from plasma samples. Samples were analyzed using NanoSight NS300. Data coutesy of Dr. Teresa Fan, University of Kentucky.
Lack of interaction between SubXTM and free phosphoproteins or phospholipids
Since bioliquids contain variety of phosphate bearing molecules including circulating cfDNA, phosphoproteins, and phospholipid monomers the question remains whether SubXTM binds to and forms aggregates with these non-exosomal phosphorylated molecules. Two “classical’ phosphoproteins – ovalbumin and casein and two abundant phospholipids - phosphatidylethanolamine and L-O-phosphoserine, have been used to answer this question. Interaction of these molecules with SubXTM were analyzed using Zetasizer Nano ZS (Malvern Instruments).
Casein molecules in aqueous solution can form micelles ranging from of 150 nm to 1 micron in diameter (Fig. 2, red line). Addition of SubXTM does not result in micelles aggregation (Fig. 2, green line), instead, SubX partly dissociate micelles back to 2-3 nm.
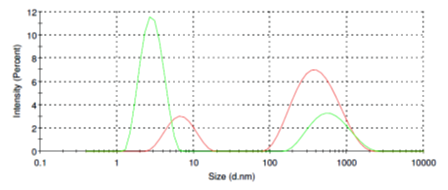
Figure 2. Size distribution of casein micelles before (red line) and after addition of SubXTM (green line).
Ovalbumin micelles in aqueous solution have more sharp distribution ranging from of 100 nm to 250 nm (Fig. 3, red line) and thus mimic extracellular vesicles pattern. Addition of SubXTM also does not result in micelles aggregation (Fig. 3, green line), instead partly dissociate micelles back to 2-3 nm.
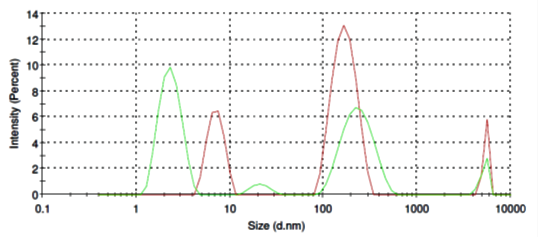
Figure 3. Size distribution of ovalbomin micelles before (red line) and after SubXTM addition (green line).
O-Phospho-L-Serine molecules form in aqueous solution micelles in a broad range from of 80 nm to 800 nm in diameter (Fig. 4, red line). Addition of SubXTM does not results in micelles aggregation (Fig. 4, green line).
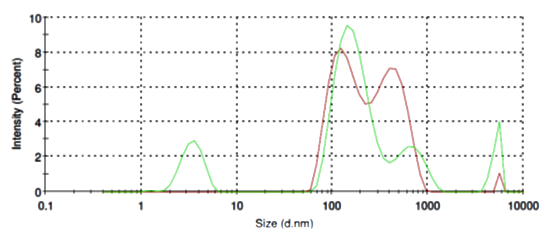
Figure 4. Size distribution of O-Phospho-L-Serine micelles before (red line) and after SubXSubXTM addition (green line).
L-Phosphatidylethanolamine molecules form in aqueous solution micelles in a range from of 400 nm to 900 nm in diameter (Fig. 5, red line). Addition of SubXTM does not results in micelles aggregation (Fig. 5, green line).
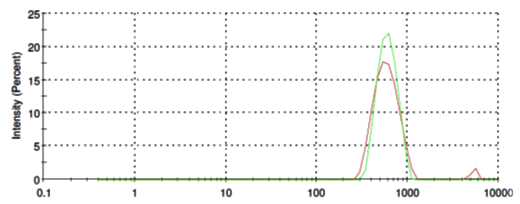
Figure 5. Size distribution of L-Phosphatidylethanolamine before (red line) and after SubXTM addition (green line).
The DNA-binding SubXTM matrix allows:
• Flexibility. Scaling up of the procedure to 50 ml of starting material or scaling down to extra-low volumes as well as adaptation of the protocol for 96- or 384-well formats
• Efficiency. Separation of cfDNA from the bulk of proteins in a single vortex-spin step without employing chaotropic agents allows you to speed-up the isolation procedure to save time and overall costs
• Adaptability. Effectively bind cell-free DNA of virtually any size ranging from 100 Kbp down to 20 bp therefore extending downstream applications
References:
Shtam T, et al. (2020) Evaluation of immune and chemical precipitation methods for plasma exosome isolation. PLOS ONE 15(11): e0242732. https://doi.org/10.1371/journal.pone.0242732
SubXTM is a trademark of Capital Biosciences, Inc.


